Intervertebral disc degeneration is accompanied by lysosomal impairment
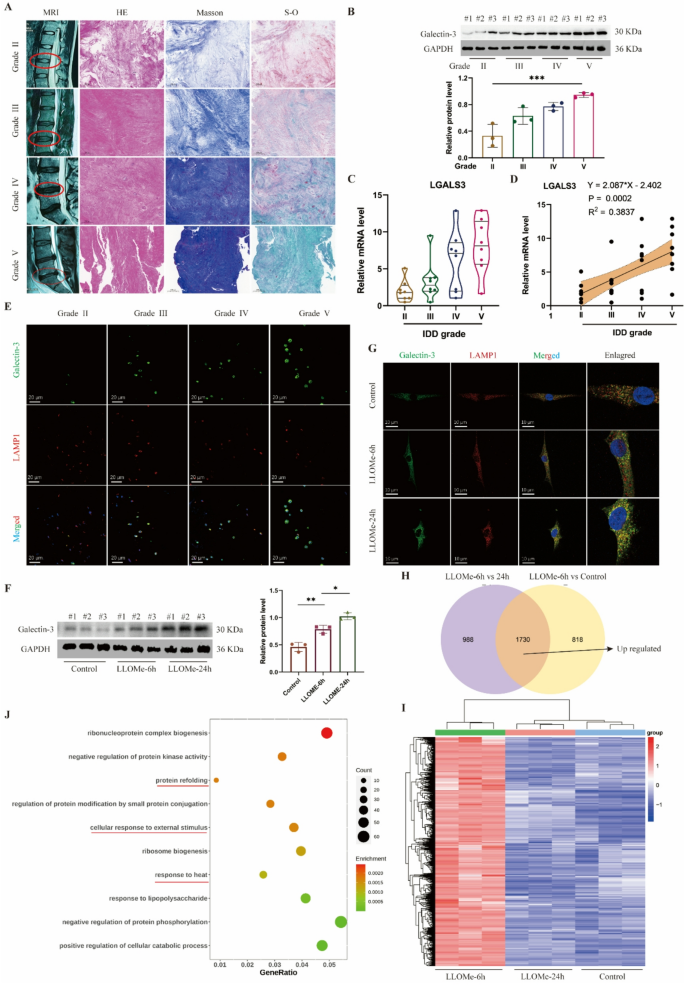
Since lysosomal dysfunction represents one of many important contributors to illness development, we sought to analyze the connection between lysosomal operate and IDD. We labeled and analyzed the degenerative grades of human intervertebral disc samples in response to the MRI pictures. Primarily based on the histological staining (Fig. 1A), samples with greater degenerative grades, akin to grades IV and V, exhibited extra densely organized tissue buildings, elevated blue depth in Masson staining, and elevated inexperienced areas in Safranin O-fast inexperienced staining. These findings collectively counsel collagen loss and elevated fibrotic adjustments in degenerated intervertebral discs (Fig. 1A). Galectin-3 serves as a marker for lysosomal impairment, and we noticed that its protein ranges progressively elevated with greater grades of disc degeneration (Fig. 1B), alongside elevated expression of the LGALS3 gene in human NP tissues (Fig. 1C). Correlation evaluation revealed a optimistic linear relationship between LGALS3 gene expression ranges and IDD grades (Fig. 1D). Moreover, immunofluorescence co-localization of galectin-3 with the lysosomal protein LAMP1 demonstrated progressively rising galectin-3 expression inside lysosomes as disc degeneration superior (Fig. 1E).
Lysosomal dysfunction is related to IDD and NP cell harm. A, Consultant MRI pictures, HE, Masson and Safranin O-fast inexperienced (S-O) staining of human intervertebral disc samples with totally different degenerative grades (Grade II, III, IV and V). B, Western blot of Galectin-3 and quantitative evaluation of protein ranges in human intervertebral disc samples. C, Relative mRNA stage of LGALS3 gene in human intervertebral disc samples. D, Correlation evaluation between the MRI grade and the expression stage of LGALS3. E, Consultant immunofluorescence of Galectin-3 and LAMP1 in human intervertebral disc samples. F, Western blot of Galectin-3 and quantitative evaluation of protein ranges in human NP cells handled with LLOMe (100 μm) at 0, 6 and 24 h. G, Consultant immunofluorescence of Galectin-3 and LAMP1 in human NP cells handled with LLOMe. H, Venn plot of up-regulated expressed genes in LLOMe-treated 6 h group. I, Heatmap of up-regulated expressed genes in LLOMe-treated 6 h group. J, GO enrichment of up-regulated expressed genes in LLOMe-treated 6 h group. Information are proven because the imply ± SD (n ≥ 3). *p < 0.05, **p < 0.01, ***p < 0.001 by a method ANOVA, or linear regression
To induce lysosomal impairment on the mobile stage, we handled human NP cells with LLOMe, a generally used lysosomal harm inducer. As LLOMe therapy length elevated, galectin-3 protein expression ranges in NP cells confirmed a progressive rise (Fig. 1F), whereas immunofluorescence outcomes confirmed elevated galectin-3 localization inside lysosomes (Fig. 1G). Concurrent with lysosomal harm, LLOMe therapy induced NP cell apoptosis, with essentially the most pronounced impact noticed on the 24-hour therapy time level (Determine S1A). To determine key regulatory molecules concerned in lysosomal restore inside NP cells, we carried out transcriptome sequencing on human NP cells handled with LLOMe for 0, 6, and 24 h (Determine S1B). Our evaluation centered on genes upregulated on the 6-hour timepoint, as these would possibly include essential early-response molecules for lysosomal harm restore. In comparison with management and 24-hour LLOMe-treated teams (Fig. 1H-I), the 6-hour therapy group confirmed 1,730 differentially upregulated genes (p-value < 0.05). GO enrichment evaluation revealed these genes had been primarily related to “protein refolding” and responses to “warmth or stimulus” (Fig. 1J), suggesting mobile self-regulative mechanisms throughout acute harm phases. KEGG pathway evaluation indicated that the upregulated genes had been associated to inflammatory signaling pathways together with TNF-α, NF-κB, and IL-17 signaling (Determine S1C). REACTOME evaluation additional demonstrated connections to mobile warmth shock responses and inflammatory reactions (Determine S1D). Collectively, our findings display a powerful affiliation between lysosomal harm and IDD. Nevertheless, the particular molecules collaborating in lysosomal restore processes in NP cells require additional elucidation.
RRAGD serves as a key molecule for sustaining lysosomal operate
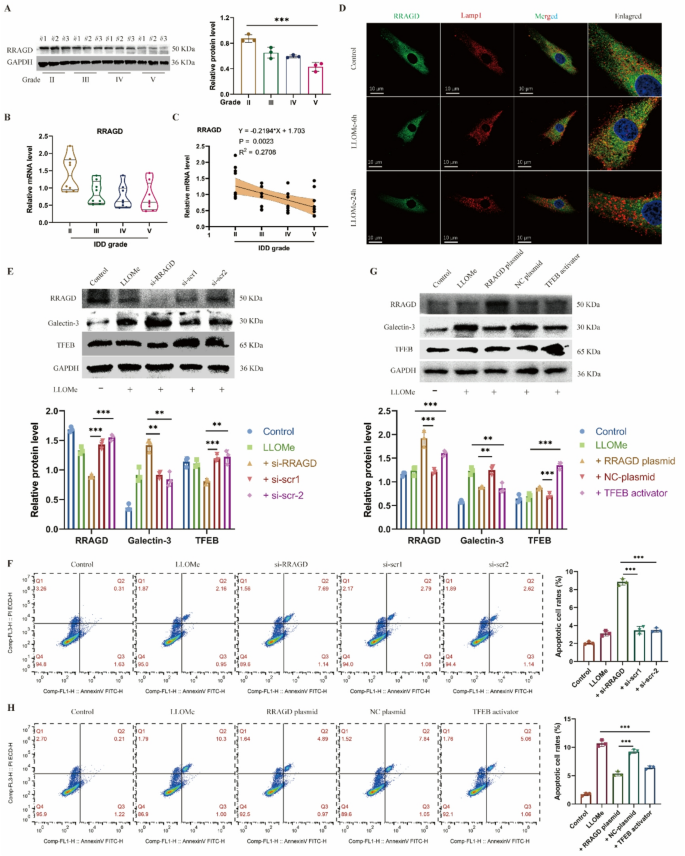
From the pool of 1,730 differentially upregulated genes, we screened these genes with operate that associated to “lysosome” and “stress response.” Amongst these, 4 genes, together with RRAGC, RRAGD, MIOS and RNF152, met the choice standards. We subsequently established a number of lysosomal harm fashions utilizing TBHP, LLOMe, and silica to induce NP cell harm. Outcomes confirmed that RRAGD expression ranges had been considerably elevated in handled teams throughout all cell fashions in comparison with controls (Determine S2A). For additional validation, we examined protein and RNA ranges of RRAGD in human NP tissues at totally different degenerative grades. Our evaluation revealed progressively reducing RRAGD protein (Fig. 2A) and mRNA expression stage (Fig. 2B) as disc degeneration superior. Correlation evaluation confirmed a detrimental linear relationship between RRAGD expression ranges and IDD degenerative grades (Fig. 2C). Notably, whereas the 6-hour LLOMe therapy group confirmed no important change in RRAGD protein ranges in comparison with controls, a considerable lower occurred after 24 h of therapy (Determine S2B). Conversely, RRAGD transcriptional ranges had been considerably elevated after 6 h of LLOMe publicity however decreased in LLOMe handled 24-hour group (Determine S2C). Immunofluorescence outcomes demonstrated RRAGD co-localization with LAMP1 on the 6-hour time level, which turned much less evident after 24 h (Fig. 2D). These findings counsel that whereas RRAGD gene transcription will increase considerably throughout early-stage stress, its expression markedly declines throughout later phases of harm.
RRAGD maintains the lysosomal operate in human NP cells. A, Western blot of RRAGD and quantitative evaluation of protein ranges in human intervertebral disc samples with totally different degenerative grades (Grade II, III, IV and V). B, Relative mRNA stage of RRAGD gene in human intervertebral disc samples. C, Correlation evaluation between the MRI grade and the expression stage of RRAGD. D, Consultant immunofluorescence of RRAGD and LAMP1 in human NP cells handled with LLOMe. E, Western blot of RRAGD, Galectin-3 and TFEB and quantitative evaluation of protein ranges in human NP cells transfected with RRAGD siRNA or scrambled siRNA. F, Stream cytometry pictures of Annexin V/PI and quantification of apoptotic cell price in respective teams. G, Western blot of RRAGD, Galectin-3 and TFEB and quantitative evaluation of protein ranges in human NP cells transfected with RRAGD plasmid or NC plasmid or handled with TFEB activator. H, Stream cytometry pictures of Annexin V/PI and quantification of apoptotic cell price in respective teams. Information are proven because the imply ± SD (n ≥ 3). *p < 0.05, **p < 0.01, ***p < 0.001 by pupil’s t take a look at, a method ANOVA, or linear regression
To additional examine RRAGD operate, we employed siRNA to inhibit the expression of RRAGD. Following therapy of RRAGD-knockdown NP cells with LLOMe, galectin-3 confirmed elevated expression whereas TFEB displayed lowered ranges, indicating the compromised lysosomal biogenesis (Fig. 2E). Cell viability assays indicated considerably decreased NP cell exercise after RRAGD interference (Determine S2D). Furthermore, RRAGD knockdown exacerbated apoptosis in LLOMe-treated NP cell (Fig. 2F). Conversely, transfection with RRAGD-overexpressing plasmids elevated each RRAGD and TFEB expression in NP cells (Fig. 2G). The RRAGD overexpression group exhibited considerably a decrease stage of galectin-3 in comparison with overexpression management, with results akin to TFEB activator therapy (Fig. 2G). Notably, RRAGD overexpression promoted restoration of cell viability (Determine S2F) and lowered cell apoptosis in comparison with LLOMe therapy alone (Fig. 2H). In abstract, RRAGD performs a vital position in sustaining regular lysosomal operate, whereas its deficiency exacerbates lysosomal harm in IDD.
The Fc-TRIM21 system gives an environment friendly platform for EV protein loading
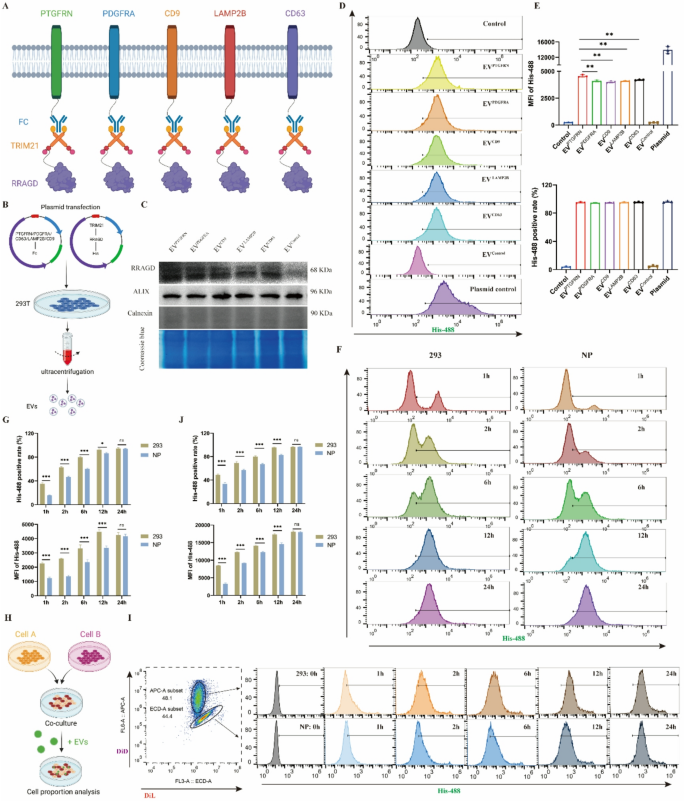
The PRYSPRY area of the Fc fragment demonstrates robust binding to its cytoplasmic receptor TRIM21. We deliberate to make the most of this Fc-TRIM21 interplay to realize focused RRAGD loading and transport inside EVs, finally aiming for lysosomal harm restore. Initially, we evaluated a number of candidate EV membrane scaffold proteins together with PTGFRN, PDGFR, LAMP2B, CD9, and CD63 to determine essentially the most appropriate platform (Fig. 3A). Co-transfection of EV scaffold protein and RRAGD plasmids into 293T cells enabled the manufacturing of engineered EVs (Fig. 3B). The outcomes confirmed detectable RRAGD in all scaffold-based engineered EVs, although expression ranges diversified (Fig. 3C). All engineered EV preparations had been detrimental for the endoplasmic reticulum protein calnexin, indicating the purity of EVs. Coomassie blue staining revealed typically related protein composition patterns throughout various kinds of engineered EV (Fig. 3C). When equal quantities of engineered EVs had been administered to NP cells, the PTGFRN group confirmed the best exogenous RRAGD stage (Determine S3A). Utilizing fluorescent His-tag to trace RRAGD-containing EV uptake, we noticed inexperienced fluorescent alerts in all engineered EV therapy teams, with the PTGFRN group exhibiting the strongest depth (Determine S3B). The stream cytometry evaluation confirmed that over 95% of NP cells internalized the engineered EVs. In comparison with controls, the engineered EV-treated group confirmed considerably elevated imply fluorescent depth (MFI), with PTGFRN-EVs demonstrating the best MFI (Fig. 3D-E). Primarily based on these outcomes, PTGFRN was chosen because the optimum scaffold protein for EV-based supply system.
Development of EV-based protein loading system. A, Schematic diagram of Fc/TRIM21 system to ship RRAGD protein primarily based on totally different EV scaffold proteins. B, Workflow of isolating engineered EVs. C, Western blot of RRAGD, Alix and Calnexin in respective engineered EVs and Coomassie blue staining. D, Stream cytometry pictures of His-488-labeled engineered EVs incubated with human NP cells. E, Imply fluorescent depth (MFI) of His-488 and His-488 optimistic cell price in respective teams. F, Stream cytometry pictures of His-488-labeled PTGFRN-engineered EVs incubated with human NP cells or 293T cells at 1, 2, 6, 12, and 24 h. G, MFI of His-488 and His-488 optimistic cell price in respective teams. H, Workflow of cell direct tradition mannequin building and cell labeling. I, Stream cytometry pictures of His-488-labeled PTGFRN-engineered EVs incubated with DiL-labeled NP cells or DID-labeled 293T cells at 0, 1, 2, 6, 12, and 24 h. J, MFI of His-488 and His-488 optimistic cell price in respective teams. Information are proven because the imply ± SD (n ≥ 3). *p < 0.05, **p < 0.01, ***p < 0.001, ns (not important) by a method or two approach ANOVA
Utilizing PTGFRN because the scaffold protein and leveraging the Fc-TRIM21 binding platform, we constructed RRAGD-delivering engineered EVs. We additional investigated potential variations in EV uptake effectivity between totally different cell sorts. Equal quantities of engineered EVs had been administered to human NP cells and 293T cells, and EV uptake was monitored. Outcomes confirmed rising proportions of EV-positive cells over time (Fig. 3F-G), although NP cells reached the height proportion extra slowly than 293T cells. To eradicate potential tradition environmental results on uptake effectivity, we established a direct coculture mannequin with NP cells and 293T cells at a 1:1 ratio (Fig. 3H). Utilizing totally different fluorescent dyes to label NP cells and 293T cells, we analyzed intracellular fluorescence depth adjustments following engineered EV therapy at numerous time factors. Each NP cells and 293T cells confirmed rising MFI and EV-positive cell proportions over time. Nevertheless, 293T cells achieved each most EV-positive cell share and highest MFI sooner than NP cells (Fig. 3I-J). These findings point out that NP cells exhibit decrease EV uptake effectivity in comparison with 293T cells, which can affect therapeutic effectiveness of engineered EVs in NP cells.
CAP-engineered EVs possess NP cell-targeting functionality
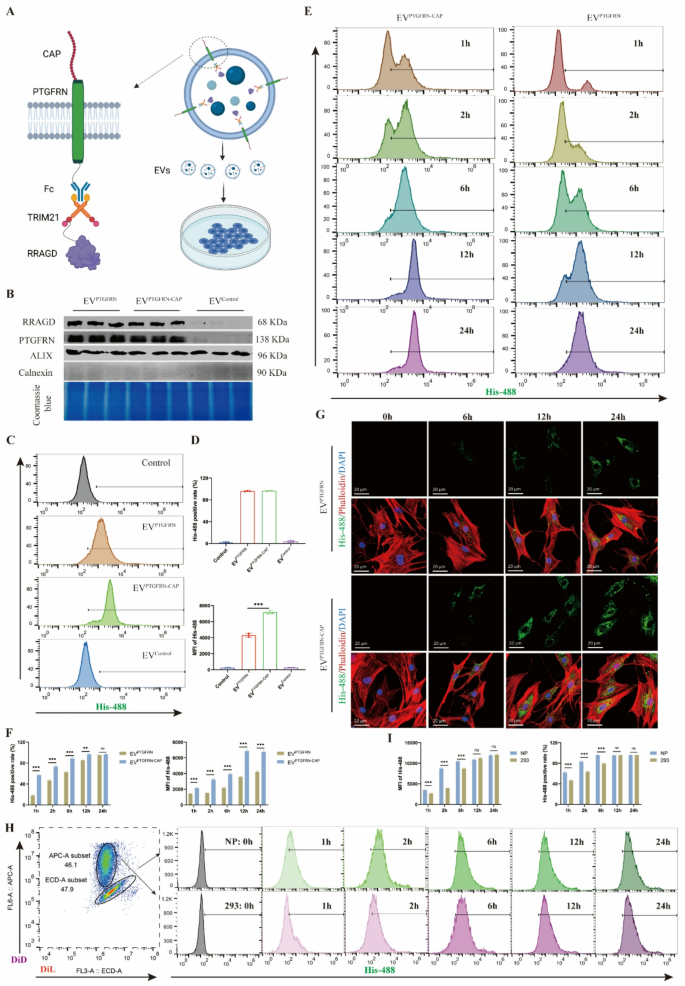
To enhance EV uptake effectivity in human NP cells, we optimized the EV membrane floor to reinforce cell concentrating on. CAP peptide, a cartilage-affinity peptide with positive binding to NP cells, was fused to the extracellular area of PTGFRN (Fig. 4A), creating an engineered EVs (EVPTGFRN−CAP). The outcomes confirmed that CAP peptide expression didn’t have an effect on bodily properties of engineered EVs, together with their morphology and dimension distribution (Determine S4A). Comparability between EVPTGFRN and EVPTGFRN−CAP revealed no important variations in RRAGD and PTGFRN protein ranges, indicating that CAP modification makes no impact on EV protein loading. The Coomassie blue staining additionally indicated minimal affect of CAP peptide incorporation on EV protein composition (Fig. 4B). When handled with equal quantities of EVPTGFRN−CAP and EVPTGFRN for an identical durations, the NP cells in EVPTGFRN−CAP group confirmed considerably greater stage of exogenous RRAGD (Determine S4B). To additional analyze the variations of EV uptake effectivity, we used fluorescent His-tagged RRAGD to trace EV internalization in human NP cells. Each EVPTGFRN−CAP and EVPTGFRN teams may detect inexperienced fluorescence alerts, with greater MFI within the CAP-modified EV group (Determine S4C). The stream cytometry outcomes confirmed comparable proportions of EV-positive cells between two teams after 24-hour EV therapy, however EVPTGFRN−CAP exhibited greater MFI of His-488 sign (Fig. 4C-D). These outcomes point out that NP cells exhibit enhanced uptake effectivity for CAP-optimized engineered EVs.
Engineering EVs with CAP peptide to acquire NP cell concentrating on. A, Schematic diagram of CAP-modified engineered EVs primarily based on Fc/TRIM21 system. B, Western blot of RRAGD, PTGFRN, Alix and Calnexin in respective engineered EVs and Coomassie blue staining. C, Stream cytometry pictures of His-488-labeled engineered EVs incubated with human NP cells. D, MFI of His-488 and His-488 optimistic cell price in respective teams. E, Stream cytometry pictures of His-488-labeled EVPTGFRN−CAP or EVPTGFRN incubated with human NP cells at 1, 2, 6, 12, and 24 h. F, MFI of His-488 and His-488 optimistic cell price in two teams. G, Consultant immunofluorescent pictures of His-488-labeled EVPTGFRN−CAP or EVPTGFRN incubated with human NP cells. Phalloidin for actin and DAPI for nucleus. H, Stream cytometry pictures of His-488-labeled EVPTGFRN−CAP incubated with DiL-labeled NP cells or DID-labeled 293T cells at 0, 1, 2, 6, 12, and 24 h. I, MFI of His-488 and His-488 optimistic cell price in respective teams. Information are proven because the imply ± SD (n ≥ 3). **p < 0.01, ***p < 0.001, ns (not important) by a method or two approach ANOVA
Prolonged time-course experiments revealed rising proportions of EV-positive cells and MFI over time within the EVPTGFRN−CAP and EVPTGFRN group. Nevertheless, the EVPTGFRN−CAP group reached peak values sooner than EVPTGFRN and achieved greater most MFI (Fig. 4E-F). Immunofluorescence staining additional confirmed persistently greater MFI within the EVPTGFRN−CAP group throughout all time factors (Fig. 4G, S4D). In addition to, coculture experiments had been carried out to check the uptake of EVPTGFRN−CAP by NP cell and 293T cell. These experiments demonstrated that each cell sorts exhibited elevated MFI and EV-positive cell proportions over time. Notably, NP cells achieved peak MFI quickly throughout coculture, reaching most EV-positive cell share and peak MFI sooner than 293T cells (Fig. 4H-I). Moreover, immunofluorescence demonstrated colocalization of internalized engineered EVs with lysosomal membrane protein LAMP1 (Determine S4E), suggesting the environment friendly supply of EVs to NP cell lysosomes. In conclusion, these findings display that CAP peptide modification confers NP cell concentrating on functionality to engineered EVs and considerably improves the EV uptake effectivity in NP cells.
Therapeutic results of engineered EVs on NP cells in vitro
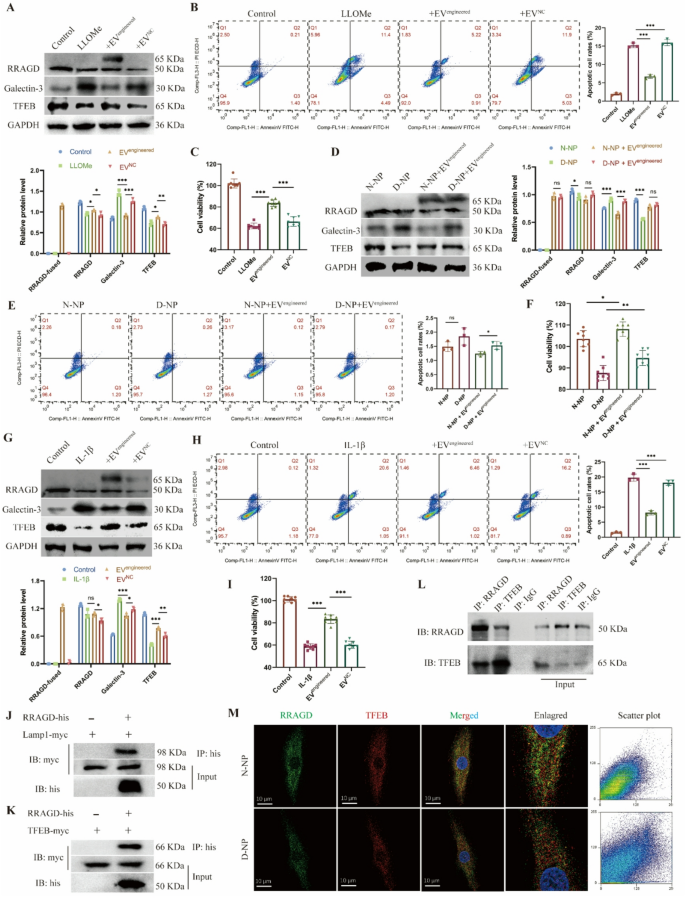
Primarily based on earlier outcomes, we constructed engineered EVs expressing CAP peptide and delivering RRAGD by way of the PTGFRN-Fc-TRIM21 platform, then evaluated the consequences in NP cell harm fashions. Within the LLOMe-induced harm mannequin, each endogenous and exogenous RRAGD expression had been detected within the engineered EV-treated group. In comparison with the LLOMe-treated group, the engineered EV group exhibited considerably decreased the galectin-3 expression and elevated the TFEB expression stage (Fig. 5A). Whereas LLOMe therapy induced NP cell apoptosis, engineered EV administration markedly inhibited this impact (Fig. 5B). Moreover, LLOMe therapy lowered each the common MFI of Calcein-staining reside cells and total cell viability, whereas engineered EVs partially restored cell survival (Fig. 5C, S5A-B). In extra experiments, we remoted NP cells from non-degenerated (N-NP) and degenerated (D-NP) intervertebral discs and incubated them with engineered EVs. The D-NP group displayed greater galectin-3 and decrease TFEB expression than N-NP cells. Engineered EV therapy lowered the galectin-3 stage in each teams (Fig. 5D). Below commonplace tradition situations with out exogenous stress, neither group confirmed important apoptosis (Fig. 5E) or variations in cell viability (Fig. 5F, S5C-D), confirming that engineered EVs didn’t have an effect on the baseline of cell viability. To simulate inflammatory harm, we handled NP cells with IL-1β, which markedly upregulated the galectin-3 stage and downregulated the TFEB expression. Engineered EVs partially counteracted these IL-1β-induced results (Fig. 5G). IL-1β therapy additionally triggered substantial apoptosis, whereas engineered EVs considerably mitigated this impact (Fig. 5H). Though IL-1β lowered live-cell fluorescence depth and viability, engineered EVs partially restored NP cell well being beneath inflammatory situations (Fig. 5I, S5E-F). Collectively, these findings display that engineered EVs exert therapeutic results on injured NP cells by enhancing viability and decreasing apoptosis.
Therapeutic results of engineered EVs on human NP cells. A, Western blot of RRAGD, Galectin-3, TFEB, and quantitative evaluation of protein ranges in human NP cells handled with LLOMe and engineered or management EVs. B, Stream cytometry pictures of Annexin V/PI and quantification of apoptotic cell price in respective teams. C, Cell viability assay in respective teams. D, Western blot of RRAGD, Galectin-3, TFEB, and quantitative evaluation of protein ranges in non-degenerated and degenerated human NP cells (N-NP and D-NP). E, Stream cytometry pictures of Annexin V/PI and quantification of apoptotic cell price in respective teams. F, Cell viability assay in respective teams. G, Western blot of RRAGD, Galectin-3, TFEB, and quantitative evaluation of protein ranges in human NP cells handled with IL-1β and engineered or management EVs. H, Stream cytometry pictures of Annexin V/PI and quantification of apoptotic cell price in respective teams. I, Cell viability assay in respective teams. J, Pull down assay of RRAGD-his and LAMP1-myc. Ok, Pull down assay of RRAGD-his and TFEB-myc. L, Co-immunoprecipitation assay of RRAGD and TFEB. IgG serves because the detrimental management. M, Consultant immunofluorescent pictures and co-localization scatter plot of RRAGD and TFEB in N-NP and D-NP cells. Information are proven because the imply ± SD (n ≥ 3). *p < 0.05, **p < 0.01, ***p < 0.001, ns (not important) by a method ANOVA
To additional elucidate the therapeutic mechanism of engineered EVs, we analyzed the interactions of RRAGD with lysosomal proteins. We constructed plasmids expressing tagged RRAGD, LAMP1, and TFEB, which had been transfected into 293T cells additional. Pull-down assays confirmed a binding interplay between RRAGD and LAMP1 (Fig. 5J), suggesting the localization of RRAGD to lysosomal membranes. Moreover, our outcomes additionally revealed that RRAGD interacted with TFEB (Fig. 5Ok), suggesting its position in lysosomal biogenesis regulation. Co-immunoprecipitation in NP cells beneath physiological situations confirmed the binding relationship between RRAGD and TFEB (Fig. 5L). Immunofluorescence additional demonstrated RRAGD-TFEB co-localization in each non-degenerated and degenerated NP cells (Fig. 5M). These observations counsel that RRAGD localizes to lysosomes and capabilities by way of binding with TFEB. In abstract, our engineered EVs successfully ship RRAGD to orchestrate lysosomal operate, thereby exerting therapeutic results on injured NP cells.
Therapeutic results of engineered EVs on disc degeneration in vivo
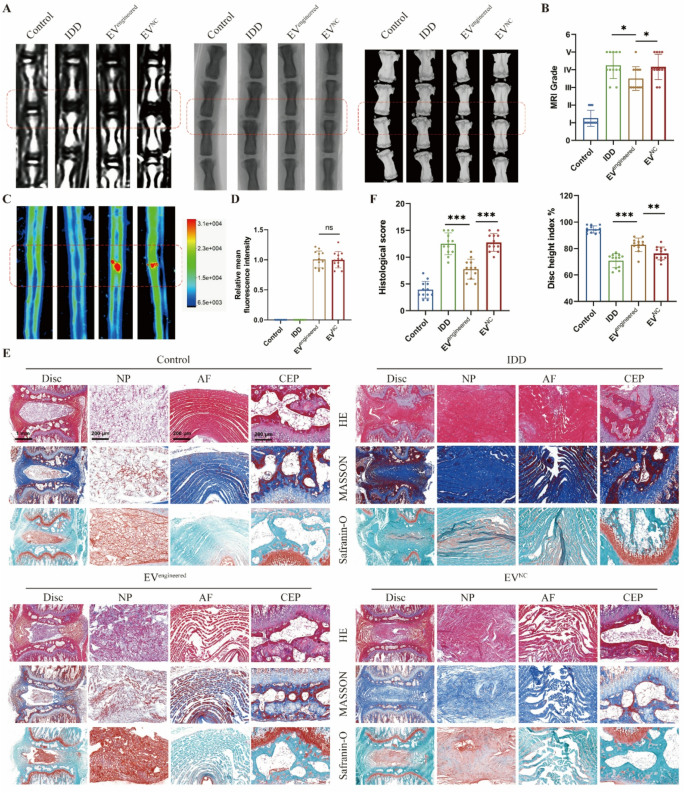
To systematically consider the therapeutic efficacy of engineered EVs in vivo, we delivered EVs into rat discs primarily based on a needle puncture-induced IDD mannequin. Disc degeneration development was comprehensively assessed by way of multimodal imaging (MRI, CT, X-ray) and histopathological evaluation. MRI quantified hydration adjustments in NP, whereas CT and X-ray detected structural alterations in intervertebral area (Fig. 6A). Degenerated discs exhibited considerably lowered hydration and collapsed intervertebral area in comparison with wholesome controls. The engineered EV group demonstrated decrease MRI grades and better Disc Top Index (DHI) versus the IDD and EV management teams (Fig. 6B). Fluorescence imaging revealed related intradiscal accumulation between engineered and management EVs (Fig. 6C-D), whereas solely engineered EVs introduced results on IDD mitigation. Histological outcomes recognized degenerative traits of discs: NP tissue atrophy, annulus fibrosus (AF) disorganization with fiber rupture, and decreased disc peak with cartilaginous endplate (CEP) degeneration. The outcomes indicated that engineered EV considerably improved histological scores in comparison with the IDD and management EV teams (Fig. 6E-F).
Engineered EVs retard the disc degeneration in vivo. A, MRI, X-ray and CT pictures of rat discs in respective teams. The rat disc was injected with engineered EVs (EVengineered) or management EVs (EVNC). B, MRI grades and Disc peak index primarily based on MRI, X-ray and CT pictures. C-D, Fluorescent pictures and relative imply fluorescence depth of rat discs in respective teams. E, HE, Masson and Safranin-O staining pictures of rat discs in respective teams. F, Histological scores primarily based on histopathological staining of rat discs in respective teams. Information are proven because the imply ± SD (n = 12). *p < 0.05, **p < 0.01, ***p < 0.001, ns (not important) by a method ANOVA
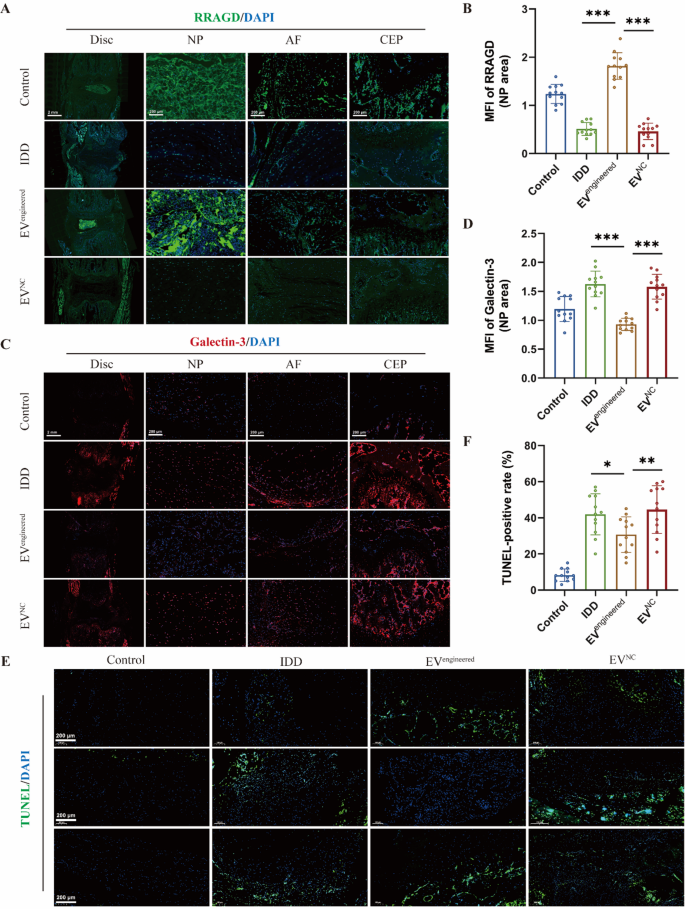
Moreover, immunofluorescence evaluation of RRAGD expression in rat discs demonstrated lowered ranges throughout disc compartments in IDD group versus management group. It revealed that engineered EVs selectively upregulated RRAGD expression in NP areas (Fig. 7A-B). It additionally confirmed that there was considerably decreased expression of galectin-3 in NP tissue of engineered EV-treated discs versus IDD and management EV group, with no important variations in AF and CEP areas (Fig. 7C-D). It demonstrated the NP-specific remedy and focused supply of engineered EVs. TUNEL staining confirmed an elevated cell apoptosis within the IDD group versus management group, whereas engineered EVs considerably lowered the speed of cell apoptosis (Fig. 7E-F). Collectively, engineered EVs successfully attenuated disc degeneration by way of focused RRAGD supply, exhibiting marked therapeutical potential for IDD.
Engineered EVs ameliorate cell loss of life throughout disc degeneration in vivo. A, Immunofluorescence of RRAGD (inexperienced) in respective teams. B, Imply fluorescence depth of RRAGD within the rat NP space in respective teams. C, Immunofluorescence of galectin-3 (crimson) in respective teams. D, Imply fluorescence depth of galectin-3 within the rat NP space in respective teams. E, TUNEL staining pictures of rat discs in respective teams. F, Quantification of TUNEL-positive cell price in respective teams. Information are proven because the imply ± SD (n = 12). *p < 0.05, **p < 0.01, ***p < 0.001 by a method ANOVA