Acquisition of nanosensor responses to intracranial tumour affected person plasma
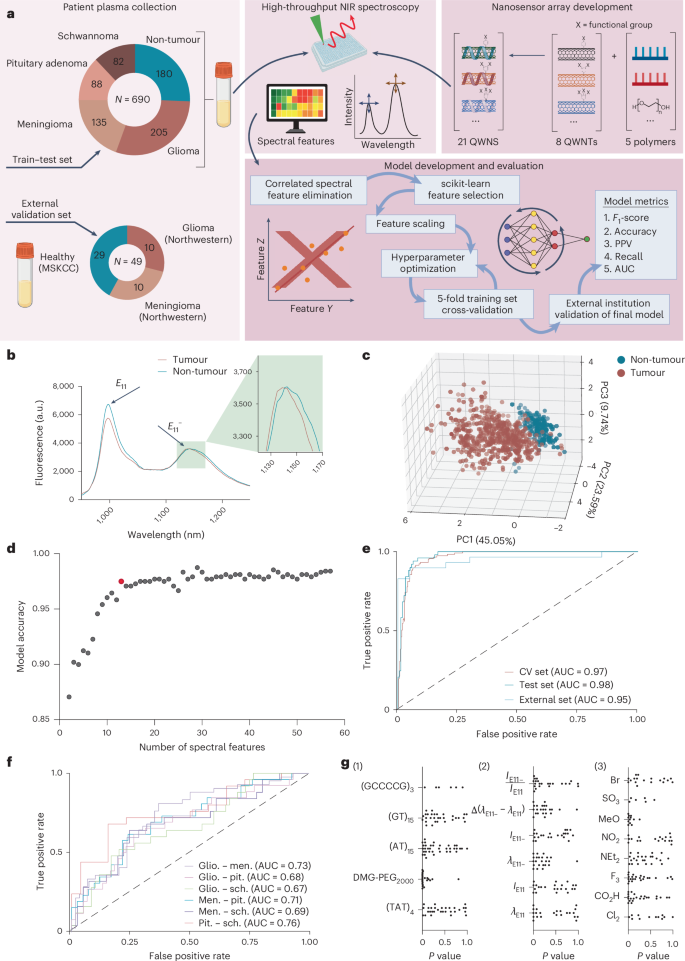
To construct our pattern dataset for mannequin growth, we collected 690 peripheral blood plasma samples from sufferers recognized on the Division of Pathology, NYU Langone Well being. These included 4 intracranial tumour cohorts representing glioma (n = 205), meningioma (n = 135), pituitary adenoma (n = 88) and schwannoma (n = 82), and 180 non-tumour samples (Fig. 1a and Supplementary Desk 1).
a, Left: schematic of diagnostic platform workflow, together with abstract of pattern courses and pattern sizes used within the research for preliminary mannequin coaching, testing and exterior establishment validation. Proper: covalently modified SWCNTs, comprised of eight useful teams (4-sulfonatebenzene-, 4-methoxybenzene-, 4-nitrobenzene-, N,N-diethylaminobenzene-, 3,4,5-trifluorobenzene-, 4-carboxybenzene-, 3,5-dichlorobenzene-, 4-bromobenzene-) have been wrapped with 5 polymers ((GT)15, (TAT)4, (GCCCCG)3, (AT)15, DMG-PEG2000) to generate an array of 21 physicochemically distinct QWNs. Centre: these QWNs have been then mixed with affected person plasma samples to probe biomolecular binding interactions utilizing high-throughput NIR spectroscopy. Backside: spectral options have been extracted and used to coach classification fashions to foretell presence and sophistication of illness. b, Consultant fluorescence emission spectra of F3-DMG-PEG2000 QWN from tumour and non-tumour samples. Spectra are the common of three technical replicates. c, PCA of QWN spectral options. Every level corresponds to a person affected person pattern, colored by pattern class. d, Mannequin efficiency as a operate of elevated variety of sensor options. The highlighted mannequin, comprising 12 spectral options, was chosen for subsequent research. e, ROC curve of cross-validation and take a look at set for tumour identification. Exterior establishment validation samples have been examined on the skilled mannequin. Dashed unity line denotes the road of no discrimination, representing the theoretical efficiency of an informationless classifier. f, ROC curve of take a look at set performances for binary classification of various tumour courses. Dashed unity line denotes the road of no discrimination. g, Evaluation of characteristic significance between tumour and non-tumour cohorts of QWN options for (1) polymer excipient (2) spectral characteristic and (3) quantum nicely covalent modification. MSKCC, Memorial Sloan Kettering Most cancers Heart; PPV, constructive predictive worth; CV, cross-validation; glio., glioma; males., meningioma; pit., pituitary adenoma; sch, schwannoma; G, guanine; C, cytosine; A, adenine; T, thymine; ({lambda }_{E11}), E11 wavelength; ({lambda }_{E11-}), E11− wavelength; ({I}_{E11}), E11 peak depth; ({I}_{E11-}), E11− peak depth; SO3, 4-sulfonatebenzene-; MeO, 4-methoxybenzene-; NO2, 4-nitrobenzene-; NEt2, N,N-diethylaminobenzene-; F3, 3,4,5-trifluorobenzene-; CO2H, 4-carboxybenzene-; Cl2, 3,5-dichlorobenzene-; Br, 4-bromobenzene-. Panel a created with BioRender.com.
To construct a sensor array with various protein coronas and optical responses to proteins, we constructed 21 QWNs comprising quantum nicely defect-modified, polymer-wrapped carbon nanotubes (Fig. 1a, Supplementary Fig. 1 and Supplementary Desk 2)14,15. We launched covalent quantum defects onto the nanotube floor utilizing a panel of aryl diazonium salts bearing substituents spanning a wide range of Hammett σ values (–0.72 to 0.78). The substituents, 4-sulfonatebenzene-, 4-methoxybenzene-, 4-nitrobenzene-, N,N-diethylaminobenzene-, 3,4,5-trifluorobenzene-, 4-carboxybenzene-, 3,5-dichlorobenzene- and 4-bromobenzene-, have been chosen to maximise range of chemical responsivities of the quantum nicely defect emission websites within the array. Every QWN generated a singular spectral profile, emitting slender fluorescence bands at 1,000 nm (intrinsic bandgap fluorescence, E11) and at ∼1,140 nm (quantum-well defect-induced emission, E11−)16 (Supplementary Fig. 1).
To additional diversify the sensitivity of QWNs within the nanosensor array, we chosen 4 single-stranded DNA sequences ((GT)15, (TAT)4, (GCCCCG)3, (AT)15) and one amphiphilic polymer (1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG2000)) (Fig. 1a) that assorted in sequence composition, construction and floor protection. The sequences (GT)15 and (TAT)4, are recognized to kind ordered floor patterns on carbon nanotubes primarily based on earlier experimental and computational research17,18, whereas (GCCCCG)3 has been predicted to undertake equally ordered conformations19, and (AT)15 is characterised by excessive floor protection20 and a disordered floor patterning. DMG-PEG2000, launched a non-DNA-based corona with distinct physicochemical properties (for instance, cost). This set was chosen to facilitate a spread of interactions with adsorbed biomolecules and to maximise sensor array range (the complete record of defect/polymer combos is given in Supplementary Desk 2).
We acquired near-infrared (NIR) fluorescence spectra by introducing every affected person pattern to wells holding every of the 21 QWNs and thrilling samples with 577-nm gentle (consultant spectra from two samples are proven in Fig. 1b). We derived six spectral options from the fluorescent emission peaks of every QWN and noticed that the QWN spectral options demonstrated various responses to the affected person samples (Supplementary Fig. 2). Principal part evaluation (PCA) of the QWN spectral responses recognized two clear clusters similar to the tumour/non-tumour pattern cohorts, with some overlap between clusters (Fig. 1c).
Classifier differentiates between intracranial tumour and non-tumour blood samples
To find out if the spectral fingerprint of QWN responses may unambiguously differentiate intracranial tumour sufferers from non-tumour plasma, we developed machine-learning fashions to discriminate between cohorts. We first used evaluation of variance (ANOVA)-based characteristic choice to find out probably the most vital spectral options from the QWN responses21. We then used the CatBoost algorithm22 to evaluate the efficiency of various characteristic combos, starting from 2 to 58 options (Supplementary Fig. 3a). To optimize the coaching mannequin, we tuned hyperparameters through Bayesian hyperparameter optimization to maximise the F1-score of 5-fold cross-validation coaching information averaged throughout folds (Supplementary Desk 3). The typical cross-validation accuracy assorted between 0.87 and 0.98 (Fig. 1d). One of the best-performing mannequin exhibited a take a look at F1-score of 0.98, an accuracy of 98%, an 88% sensitivity at 95% specificity, and an space beneath the curve (AUC) of the receiver working attribute (ROC) curve of 0.98 (Fig. 1e). Strikingly, as evidenced by the excessive total prediction accuracy, our fashions have been capable of detect tumours at a spread of WHO grades, and over 20% of our glioma cohort was comprised of WHO grade 1–2 tumours (Supplementary Fig. 4a). The statistical evaluation of age, intercourse and sophistication imbalance on mannequin efficiency confirmed negligible bias (Supplementary Fig. 4b–d and Supplementary Desk 4).
To find out the generalizability of this technique, we assembled a validation cohort of samples from two exterior establishments. For this evaluation, we synthesized a brand new set of QWNs. Utilizing the brand new batch of nanosensors, we collected spectral responses from 20 main tumour plasma samples and 29 non-tumour plasma samples from Northwestern College and the Memorial Sloan Kettering Most cancers Heart (Fig. 1a and Supplementary Desk 5). The QWN responses clustered persistently with our preliminary cohort of 690 affected person samples, confirming strong sensor responses (Supplementary Fig. 5). The efficiency of this exterior validation dataset of the beforehand skilled mannequin was akin to the interior take a look at set with an F1-score of 0.897, an accuracy of 89.8%, a specificity of 93.1% at a sensitivity of 98%, and an AUC of 0.95 (Fig. 1e), suggesting reproducible sensor synthesis and robustness of spectral responses to affected person samples
We investigated the potential of the QWN array to discriminate between intracranial tumour varieties. The QWN optical responses to plasma from sufferers harbouring the 4 intracranial tumour varieties (glioma, meningioma, pituitary adenoma and schwannoma) didn’t separate into particular person clusters through PCA (Supplementary Fig. 6a). We skilled CatBoost classification algorithms to distinguish between every pair of tumours (Fig. 1f and Supplementary Fig. 6b–g). The classifier skilled to distinguish between glioma and meningioma scored the very best among the many differentiation duties, with a weighted F1-score of 0.71, an accuracy of 71%, a precision of 0.74 and an AUC of 0.73 (Supplementary Fig. 6b). The opposite fashions carried out with accuracies ranging between 65% and 71% (Supplementary Fig. 6b–g; detailed mannequin statistics are given in Supplementary Desk 6). The detection fashions weren’t capable of robustly establish all clinically related variations, nonetheless. For instance, detection of the isocitrate dehydrogenase 1 (IDH-1) mutation standing of glioblastoma sufferers gave a take a look at set AUC of 0.51, performing no higher than likelihood (Supplementary Fig. 7).
3,4,5-F3-DMG-PEG2000 QWN drives machine prediction efficiency
We aimed to establish which QWN options have been chargeable for the disease-specific sensor responses. The DMG-PEG2000-wrapped QWNs have been statistically overrepresented within the vital characteristic set (Fig. 1g(1), ({chi }^{2}=27.34;,P < 1.7times {10}^{-7})), suggesting that the DMG-PEG2000 wrapping might facilitate the binding of disease-relevant analytes to the QWN floor. The only most necessary sensor for the classification activity was the three,4,5-trifluoro aryl functionalized nanotube with a DMG-PEG2000 wrapping, termed the three,4,5-trifluoro-DMG-PEG2000 nanosensor, which alone detected tumours with 93% accuracy and differentiated between glioma and meningioma with 64% accuracy (Supplementary Fig. 3). The response was additional improved by the addition of different sensors. The E11− wavelength spectral characteristic was usually a very powerful for mannequin growth as assessed by the scikit-learn characteristic choice toolkit (Fig. 1g(2)), in line with earlier work suggesting that the quantum nicely defects can elicit enhanced environmental sensitivity23. Correlation evaluation between spectral responses to affected person plasma samples (Supplementary Figs. 8 and 9) indicated that many spectral options transduced a considerable diploma of mutual data, highlighting the necessity for characteristic choice.
MPLB platform reveals potential biomarkers driving disease-specific QWN responses
To analyze the molecular species driving the MPLB illness response, we carried out quantitative proteomic evaluation on non-tumour plasma samples and on samples from glioblastoma and meningioma sufferers (Fig. 2a) to establish the protein corona of the one most predictive QWN, 3,4,5-trifluoro-DMG-PEG2000. After washing away weakly sure proteins from the QWN corona, we eluted the remaining proteins (Fig. 2a and Supplementary Fig. 10a,b)24 and carried out quantitative mass spectrometry analyses on three unbiased protein corona extractions for every cohort, labelled utilizing 18-plexed TMTpro tandem mass tags to allow pattern multiplexing25 (Fig. 2a and Supplementary Fig. 10a,b). PCA revealed that non-tumour and tumour pattern clusters separated, whereas glioblastoma and meningioma clusters overlapped (Supplementary Fig. 10d), mirroring the QWN optical responses (Fig. 1c and Supplementary Fig. 6a). The protein compositions of the glioblastoma and meningioma samples have been considerably correlated, with a Pearson coefficient of 0.86. This, in distinction to 0.75 for glioblastoma and non-tumour samples and 0.78 for meningioma and non-tumour samples (Supplementary Fig. 10e), suggests better similarity between the protein corona compositions of the tumour samples, and extra variations between non-tumour and tumour samples, additional recapitulating the traits in QWN mannequin efficiency.
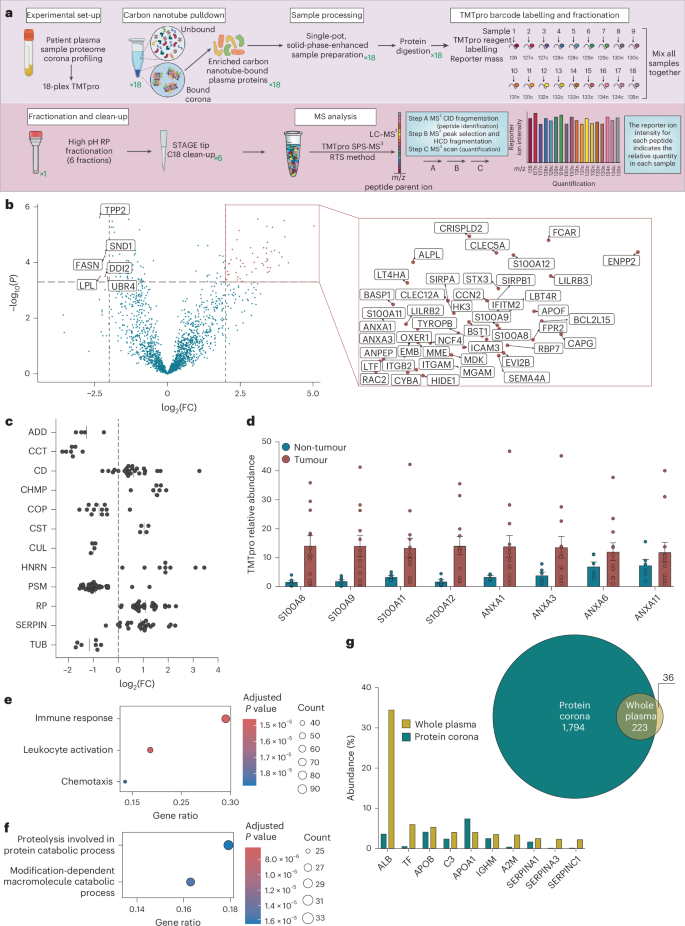
a, Schematic of QWN corona proteomics workflow. b, Volcano plot of log2(FC) versus −log10(P) between tumour (n = 12) and non-tumour (n = 6) affected person plasma protein corona extracts; protein IDs labelled with |FC| > 4 and P< 5 × 10−4 are highlighted. Samples have been analysed utilizing two-sided Welch’s t-test, adjusted for a number of comparisons at a 1% FDR. c, log2(FC) of tumour (n = 12) in comparison with non-tumour (n = 6) samples of chosen protein courses, indicating elevated and depleted expression throughout many courses of proteins. d, TMTpro relative abundance of particular proteins related to tumour samples (n = 12) in comparison with non-tumour (n = 6). Information are reported as imply ± s.e.m. e, Gene Ontology enrichment evaluation of proteins with |log2(FC)| > 1. Circle measurement signifies variety of considerably enriched proteins with useful annotation and circle color signifies statistical significance. Statistical take a look at was carried out through one-sided Fisher’s actual take a look at, with Benjamini–Hochberg correction at 5% FDR. f, Gene Ontology depletion evaluation of proteins with |log2(FC)| > 1. Circle measurement signifies variety of considerably depleted proteins with useful annotation and circle color signifies statistical significance. Statistical take a look at was carried out through one-sided Fisher’s actual take a look at, with Benjamini–Hochberg correction at 5% FDR. g, Higher proper: Venn diagram of proteins detected through LC–MS/MS in matched protein corona extraction (n = 18) and whole-plasma enter (n = 18). Decrease left: common share abundance of the ten most prevalent plasma proteins from mass spectrometry evaluation in entire plasma (n = 18) and their abundance in protein corona extractions (n = 18). TMT, tandem mass tag; RP, reverse section; STAGE, cease and go extraction; RTS, real-time search; CID, collision-induced dissociation; HCD, higher-energy collisional dissociation; ADD, adaptor protein; CCT, chaperonin-containing TCP-1; CD, cluster of differentiation; CHMP, charged multivesicular physique protein; COP, coatomer protein; CST, cystatin; CUL, cullin; HNRN, heterogeneous nuclear ribonucleoprotein; PSM, proteasome subunit; RP, ribosomal subunit; SERPIN, serine protease inhibitor; TUB, tubulin; Alb, albumin; TF, transferrin; APOB, apolipoprotein B; C3, complement C3; APOA1, apolipoprotein A1; IGHM, immunoglobulin heavy fixed μ; A2M, α-2-macroglobulin; SERPINA3, serine protease inhibitor A3; SERPINC1, serine protease inhibitor C1. Panel a created with BioRender.com.
After elution, we noticed 2,017 QWN-enriched proteins, 1,155 of which have been enriched in tumour affected person pattern proteins, as in comparison with 862 proteins enriched in non-tumour people (Fig. 2b). A full record of considerably enriched and depleted proteins is supplied in Prolonged Information Desk 1 The 5 most importantly enriched proteins within the corona of tumour sufferers have been ENPP2, LILRB3, FCAR, BCL2L15 and CAPG (Prolonged Information Desk 1). These proteins point out a protein corona composition derived from tumour samples influenced by proteins concerned in irritation26, immune modulation27,28 and lipid signalling29, launched from the TME, and by systemic inflammatory responses. The 5 most importantly depleted proteins within the corona of tumour sufferers have been TPP2, FASN, LPL, DDI2 and PDE5A, (Prolonged Information Desk 1). These proteins are concerned in numerous roles together with proteostasis30, lipid metabolism31 and RNA regulatory mechanisms32.
Gene Ontology evaluation33 revealed differentially enriched proteins within the tumour samples (|log2(FC)| > 1; FC, fold change). The proteasome 19S and 20S subunits (PSMs) have been considerably depleted on the floor of the carbon nanotube in most cancers sufferers (Fig. 2c). In distinction, the ribosomal subunits (RPs) have been persistently extra considerable in tumour samples (Fig. 2c), in line with earlier research exhibiting enrichment of RPs in glioblastomas34.
We discovered that a number of S100A proteins have been notably upregulated in tumour samples, particularly S100A8, S100A9, S100A11 and S100A12 (Fig. 2nd). These S100A proteins have been acknowledged as potential biomarkers in numerous neurological tumours35,36,37. Moreover, our evaluation indicated that ANXA1, 3, 6 and 11 have been considerably enriched within the tumour samples (Fig. 2nd), aligning with earlier analysis38. Moreover, TME and inflammatory response proteins have been recognized, together with FCAR, LILRB2/B3, SIRPA/B3 and CLEC5A/12A. The overwhelming majority of the enriched proteins (proven in Fig. 2b,c) haven’t beforehand been reported as tumour biomarkers. Determine 3e,f experiences highlighted GO organic processes of differentially enriched proteins (| log2(FC)| > 1), indicating that enriched proteins are related to immune responses, leukocyte activation and chemotaxis (Fig. 2e), whereas the depleted proteins are linked to proteolysis and catabolysis (Fig. 2f). The information recommend {that a} mixture of tumour-specific, tumour microenvironment, and systemic inflammatory and immunological proteins from tumour samples have been enriched on the QWNs (Prolonged Information Desk 1).
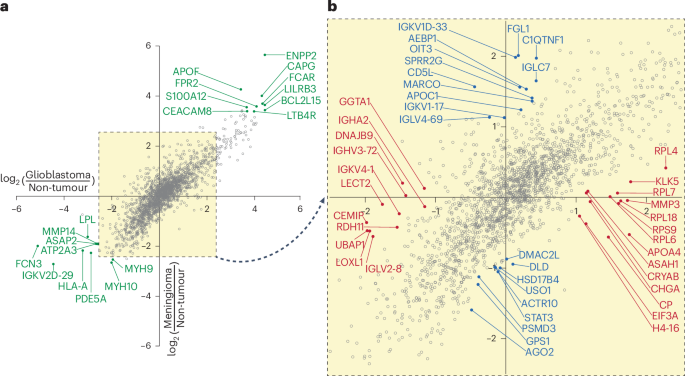
a, Plot of glioblastoma/non-tumour log2(FC) versus meningioma/non-tumour log2(FC), highlighting correlation between tumour enrichment and depletion profiles. Highlighted proteins in gentle inexperienced are the highest 10 tumour non-specifically enriched (upper-right quadrant) or depleted (lower-left quadrant), representing potential molecular markers of most cancers, agnostic to most cancers sort. Dashed yellow field with arrow signifies highlighted area of plot that’s expanded in b. b, Magnified view at x intercept and y intercept reveals potential most cancers type-specific differentially enriched protein species. Crimson, enriched (x > 0, y = 0) or depleted (x < 0, y = 0) in glioblastoma samples in contrast with non-tumour samples; blue, enriched (x = 0, y > 0) or depleted (x = 0, y < 0) in meningioma samples in contrast with non-tumour samples.
Protein corona evaluation facilitates the detection of low-abundance biomarkers
To judge the extent to which our QWN protein-enrichment method helps the detection of low-abundance proteins, we analysed the variations within the proteomic information supplied by QWN-eluted plasma in comparison with whole-plasma samples. We carried out an 18-plex TMTpro experiment utilizing matched affected person plasma samples (non-tumour, glioblastoma and meningioma) and in contrast it to the protein corona extracts detected from the QWN. In distinction with the two,017 proteins recognized from the QWN, we may reliably quantify solely 259 proteins within the undepleted plasma extracts (Fig. 2g, inset). Statistical evaluation revealed that a number of of the proteins enriched in tumours, together with S100A8, S100A9 and ANXA1, have been detectable utilizing bulk plasma enter (Supplementary Fig. 11), however the overwhelming majority of tumour-enriched proteins recognized from QWN elution weren’t quantified within the bulk plasma enter. In matched samples, albumin was over 10-fold depleted, and transferrin was 17-fold depleted within the protein corona in contrast with entire plasma (Fig. 2g). Of the highest 10 most considerable proteins within the undepleted plasma samples, solely APOA1 confirmed a relative enrichment (1.4-fold) within the protein corona in contrast with the entire plasma. This discovering is in line with the better repertoire of protein varieties detected in protein corona (n = 2,017) in comparison with undepleted plasma (n = 259).
Identification of most cancers type-specific protein enrichment
Subsequent, we targeted on figuring out QWN-eluted proteins that exhibited particular differential expression between both glioblastoma and non-tumour samples or between meningioma and non-tumour samples. There was substantial collinearity between the protein enrichment profiles between non-tumour/glioblastoma and non-tumour/meningioma samples (Supplementary Fig. 10e), suggesting that a big part of the variation was influenced by protein-binding interactions not particular to tumour class (Fig. 3a). By evaluating alongside the x and y intercepts, we recognized most cancers type-specific variations within the composition of the QWN protein corona (Fig. 3b).
Alongside the x intercept, we recognized a number of glioblastoma-specific protein biomarkers within the composition of the QWN protein corona, compared to non-tumour samples (Fig. 3b and Prolonged Information Desk 2). Identified biomarkers, together with matrix metalloproteinase 3 (MMP3)39, apolipoprotein A4 (APOA4)40 and several other RPs (RPL4, RPL7, RPL18, RPS9, RPL6)34, have been enriched 2- to three.5-fold on the protein corona of the glioblastoma samples, whereas they weren’t differentially enriched within the meningioma protein corona samples. Different differentially enriched proteins weren’t beforehand acknowledged as implicated in glioblastoma, however are variously concerned in extracellular matrix (ECM) remodelling, supporting progress, enhancing stress resistance and response, or are associated to immune operate pathways (Prolonged Information Desk 2).
Our evaluation alongside the y intercept revealed meningioma-enriched proteins in comparison with non-tumour samples, all unreported as meningioma biomarkers (Fig. 3b and Prolonged Information Desk 3). Enrichment of immunological proteins (HLA-B, MARCO, CD5L)41,42,43 and ECM elements (AEBP1)44 are in line with the immune and structural composition of meningiomas. Moreover, the enrichment of lipid metabolism proteins (APOC1, C1QTNF1) helps the present literature on adjustments in meningioma lipid metabolism45. Curiously, most of the proteins recognized all through our proteomic analyses weren’t usually secreted proteins (Prolonged Information Desk 3), which means that the noticed proteins outcome from mobile fragments launched from the tumour into the bloodstream.
Identification of differentially enriched proteins between glioblastoma and meningioma sufferers driving the prediction mannequin
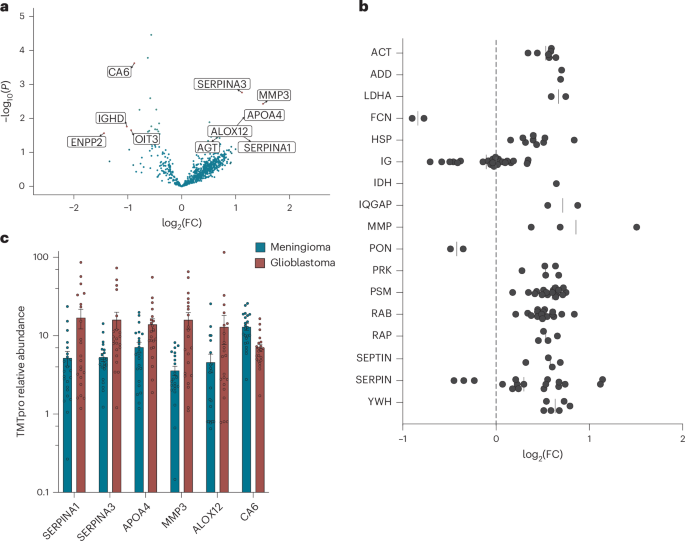
We assessed the protein-binding profiles doubtlessly undergirding the mannequin efficiency of our glioblastoma and meningioma classifier (AUC, 0.73; Fig. 1f). We built-in findings from three TMTpro protein corona extraction experiments to boost the statistical sign and establish potential proteins contributing to mannequin efficiency (Fig. 1f and Supplementary Fig. 6b). After combining the protein quantification outcomes from the three experiments, we noticed a complete of 817 distinctive QWN-enriched distinctive proteins throughout all three experiments. Of those, 645 have been enriched in glioblastoma affected person samples, whereas 172 proteins have been enriched in meningioma affected person samples (Fig. 4a). We discovered that numerous protein courses have been differentially sure to the sensors throughout teams, together with many members of the septin household, heat-shock proteins and the protein kinase C-related household. These proteins serve various roles in ECM and structural remodelling, and in immunological features (Fig. 4b). Our outcomes confirmed that a number of serpins, together with SERPINA1, SERPINA3 and AGT, have been comparatively enriched within the protein corona of glioblastoma sufferers (Fig. 4a,c). Notably, SERPINA3 has beforehand been recognized as a diagnostic biomarker of glioblastoma40,46. Moreover, MMP339 and APOA440 have been reported as upregulated proteins in glioblastoma sufferers, which aligns with our findings (Fig. 4c)40,46. We discovered that ENPP2, CA6, OIT3 and IGHD have been comparatively enriched within the protein corona blood plasma of meningioma sufferers, enjoying roles in lipid processing and immune responses (Fig. 4a,c). Importantly, the diploma of overlap with the literature additional means that each recognized and unknown protein illness biomarkers influenced the sensor responses.
a, Volcano plot of log2(FC) versus −log10(P) between glioblastoma (n = 22) and meningioma (n = 23) samples. Samples have been analysed utilizing two-sided Welch’s t-test, adjusted for a number of comparisons at a 1% FDR. b, log2(FC) of glioblastoma (n = 22) in contrast with meningioma (n = 23) corona samples of chosen protein courses, indicating elevated and depleted expression throughout many courses of proteins. c, Relative protein abundance between glioblastoma (n = 22) and meningioma (n = 23) samples of chosen differentially enriched corona proteins. Information are reported as imply ± s.e.m. ACT, actin; ADD, adducin; LDHA, lactate dehydrogenase a; FCN, ficolin; HSP, heat-shock protein; IG, immunoglobulin; IDH, isocitrate dehydrogenase; IQGAP, iq motif-containing GTPase-activating protein; MMP, matrix metalloproteinase; PON, paraoxonase; PRK, protein kinase; PSM, proteasome subunit; SERPIN, serine protease inhibitor; YWH, 14-3-3 protein.
Differentially enriched QWN corona proteins generate quantitative sensor responses
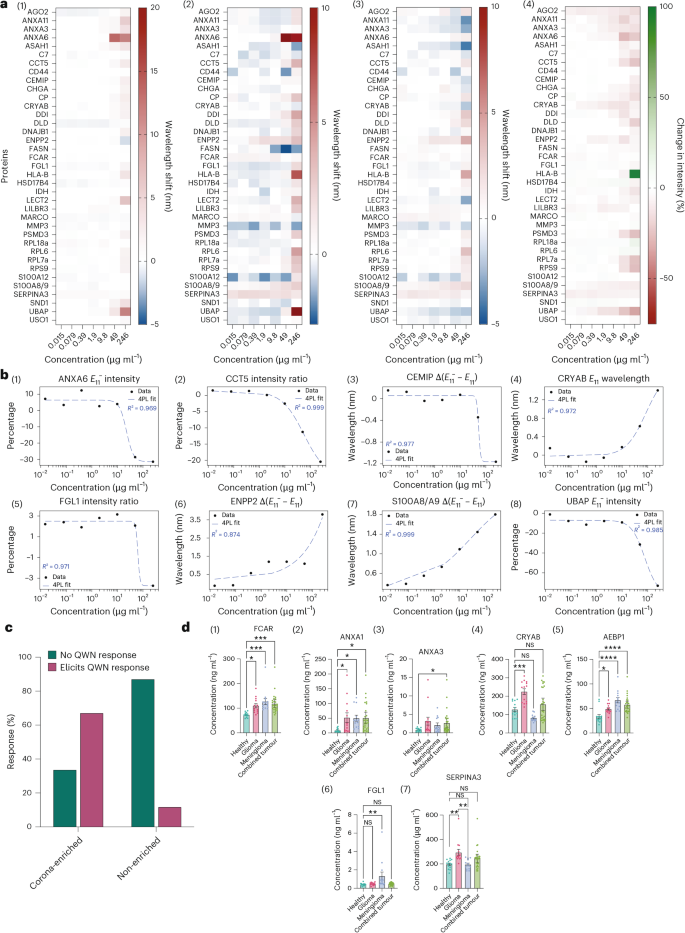
We carried out protein titration experiments to evaluate the spectral responses of three,4,5-trifluoro-DMG-PEG2000 to 36 of the recognized disease- and corona-enriched proteins, and to a set of twenty-two proteins that weren’t enriched within the protein corona, at concentrations starting from 246 to 0.015 μg ml−1 in 5-fold dilutions utilizing interferent pooled wholesome plasma (20%) or phosphate-buffered saline (PBS) (Supplementary Desk 7). The QWNs exhibited delicate and quantitative spectral responses to sure proteins (Fig. 5a(1)–(4), Supplementary Figs. 12 and 13). Within the absence of plasma interferent (that’s, in PBS), the magnitude of the sensor response at most protein focus was elevated ((bar{{lambda }_{{E}_{11}^{-}}}:4.41;{mathrm{{nm}}})) in comparison with the protein interferent situation ((bar{{lambda }_{{E}_{11}^{-}}}:2.27 ;{{mathrm{nm}}})). Nonetheless, total response traits between PBS and interferent circumstances have been broadly constant, significantly between ratiometric spectral parameters (Fig. 5a(4) and Supplementary Fig. 13(6)). We fitted four-parameter log-logistic (4PL) fashions to the dose–response curves and located that most of the sensor responses to titrated biomarkers have been well-described (R2 = 0.87–0.99). Determine 5b highlights robust quantitative QWN responses to among the most extremely enriched proteins (Figs. 2a and 3b), indicating that binding interactions between these enriched proteins and QWNs can elicit substantial spectral adjustments to QWN emission.
a, Warmth maps of F3-DMG-PEG2000 spectral responses to 36 QWN-identified non-specific most cancers and tumour-specific potential biomarkers: (1) change in E11 wavelength; (2) change in E11− wavelength; (3) change in E11− − E11 wavelength; (4) change in peak depth ratio. b, Dose–response curves to pick candidate biomarkers within the presence of plasma interferent with four-parameter log-logistic mannequin match overlaid (blue dashed line). Proteins have been chosen on the idea of quantitative logistic spectral responses (R2 > 0.7) and significance in QWN corona proteomics fold-enrichment: (1) ANXA6 E11− depth; (2) CCT5 depth ratio; (3) CEMIP ΔE; (4) CRYAB E11 wavelength; (5) FGL1 depth ratio; (6) ENPP2 ΔE; (7) S100A8/A9 ΔE; (8) UBAP E11− depth. c, Quantification of protein responses for 58 proteins, comprised of 36 QWN binders, recognized through proteomics, and 22 non-binders, which weren’t detected or not enriched in protein corona analyses. Responders are proteins that elicited a response in a number of spectral options as decided by R2 of match >0.7 and response at maximal focus >1 nm or 20% change in depth. d, ELISA protein quantification of 39 whole-plasma samples (n = 13 wholesome donor, 13 glioblastoma, 13 meningioma). Information are reported as imply ± s.e.m. Samples have been analysed utilizing two-sided Welch’s t-test, adjusted for a number of comparisons at a 5% FDR. *P < 0.05; **P < 0.01; ***P < 0.001, ***P < 0.0001, ****P < 0.00001. (1) ANXA1; (2) ANXA3; (3) FCAR; (4) CRYAB; (5) AEBP1; (6) FGL1; (7) SERPINA3. int, depth; 4PL, four-parameter log-logistic mannequin; NS, non-significant.
We categorized every protein as eliciting a QWN response or not primarily based on two standards: (1) the goodness of match of dose–response curves (R2 > 0.7) and (2) the response at 246 μg ml−1 (>1-nm wavelength shift or >20% depth change) within the presence of plasma interferent. The proportion of corona-enriched and non-enriched proteins elicited a response in a number of spectral options (Fig. 5c) and the enriched proteins (67%; n = 36) have been considerably extra possible (Fisher’s actual take a look at; odds ratio, 0.143; P < 0.0041) to elicit a quantitative sensor response in comparison with the non-enriched proteins (14%; n = 22). This evaluation signifies that proteins enriched within the protein corona elicit quantitative QWN responses, generate attenuated however not abrogated responses within the presence of plasma interferent, and usually tend to elicit sensor responses than proteins not enriched within the protein corona.
ELISAs validate protein corona enrichment traits
We investigated whether or not the enrichment of newly recognized candidate biomarkers from the QWN protein corona aligned with the protein enrichment in entire plasma. We carried out enzyme-linked immunosorbent assays (ELISAs) to quantify 11 QWN-enriched proteins (Supplementary Desk 8) in 39 affected person samples (13 non-tumour, 13 glioblastoma and 13 meningioma). We noticed a powerful correlation between the QWN corona proteomics identification of differentially enriched corona proteins and whole-plasma ELISA detection. Particularly, differentially enriched tumour non-specific proteins, together with ANXA1, ANXA3 and FCAR (Fig. 5d(1)–(3)) confirmed elevated presence in each tumours in contrast with wholesome samples (Fig. 2b). AEBP1 and FGL1 have been detected in considerably larger portions in meningioma samples in contrast with each wholesome and glioblastoma samples (Fig. 5d(4),(5)), which aligns with our proteomics outcomes (Fig. 3b). CRYAB demonstrated constant enrichment (Fig. 5d(6)) in glioblastoma samples in contrast with wholesome and meningioma samples, in line with Fig. 3b. Lastly, SERPINA3 was enriched in glioblastoma sufferers relative to meningioma sufferers (Fig. 5d(7)), in line with our direct glioblastoma–meningioma comparability (Fig. 4a). General, 7 of the 11 QWN-enriched proteins examined achieved statistical significance for both or each tumour varieties within the whole-plasma samples (Fig. 5d and Supplementary Fig. 14).